Crop Anomaly Detection Tool: How One Agronomist Used n8n, Qdrant & Voyage AI To Catch The Unknown
On a hot afternoon at the edge of a sprawling farm, Priya, an agronomist and data lead for a precision agriculture startup, stared at a dashboard that would not stop blinking red.
Her team had just rolled out a new field scouting app. Farmers were sending in thousands of crop photos every week. The idea was simple: classify each image into known crop types, then flag anything unusual for closer inspection. In practice, it was chaos.
Some images were mislabeled. Others contained pests and diseases that the training dataset had never seen. A few were not even farm crops at all. Her models tried to force every image into one of the known classes, and the result was a dangerous mix of overconfident predictions and missed anomalies.
What she needed was not just better classification. She needed a production-ready crop anomaly detection system that could say, with confidence, “this image does not belong to any known crop class.”
That search led her to an n8n workflow template built on Qdrant and Voyage AI.
The Problem: When “Unknown” Is The Most Important Class
Priya’s team managed a 29-class crop image dataset, covering staples and cash crops such as:
- pearl_millet (bajra), tobacco-plant, cherry, cotton, banana, cucumber, maize, wheat
- clove, jowar, olive-tree, soyabean, coffee-plant, rice, lemon, mustard-oil
- vigna-radiati (mung), coconut, gram, pineapple, sugarcane, sunflower, chilli
- fox_nut (makhana), jute, papaya, tea, cardamom, almond
Their classifiers were decent at telling wheat from maize or cotton from tobacco. What they could not do reliably was answer a different, far more critical question:
- Is this photo a completely unknown plant species that slipped into the frame?
- Does it show a pest, disease, or damage pattern that was not part of the original training data?
- Is the image itself mislabeled or corrupted in the dataset?
In other words, they needed a system that could say, “this looks unlike anything we know,” instead of bending reality to fit a known label. That is where anomaly detection came in.
Discovering a Different Approach: Medoids, Embeddings and Vector Search
During a late-night search through developer forums and automation communities, Priya found a reference to an n8n-based workflow that combined Voyage AI multimodal embeddings with a Qdrant vector database.
The idea clicked immediately.
Instead of forcing each new image through a classification model, the workflow would:
- Turn the input image into a high dimensional embedding vector using a Voyage AI multimodal model.
- Compare that embedding against cluster centers (medoids) for each crop class stored in a Qdrant collection.
- Use per-cluster similarity thresholds to decide whether the image was close enough to any known crop type.
- If no medoid passed its threshold, mark the image as anomalous, likely a new or undefined crop.
This was not just classification. It was a compact and explainable anomaly detection pipeline, and it could be automated end to end inside n8n.
Setting the Stage: Building the Crop Collection in Qdrant
Before Priya could rely on the anomaly detector, she needed a solid foundation in Qdrant. That meant preparing the dataset and computing medoids for each crop class.
Step 1 – Preparing the dataset
Her team started with a public dataset, similar to the Kaggle “agricultural crops” dataset. They:
- Downloaded all crop images.
- Uploaded them to a cloud storage bucket accessible from their pipelines.
- Kept labels and metadata intact for each image.
Step 2 – Populating Qdrant with embeddings
Using a separate n8n pipeline, they:
- Generated Voyage AI embeddings for each image.
- Uploaded these embeddings, along with labels and metadata, into a Qdrant collection.
- Used a free tier Qdrant Cloud cluster for initial experiments.
At this stage, Qdrant stored all points, but Priya knew the anomaly detection workflow would work best with medoids and cluster thresholds per crop class.
Step 3 – Computing medoids and thresholds
A dedicated “medoids setup” pipeline calculated, for each crop class:
- The medoid point that best represented the class center.
- A cluster threshold, a similarity score cutoff that a new image must reach to be considered part of that class.
In Qdrant, each medoid point received two crucial payload fields:
is_medoid(boolean) – marks the point as the medoid/centroid of its cluster.is_medoid_cluster_threshold(float) – the minimum similarity score required for a match to that cluster.
This design meant that each crop class could have its own tolerance for variability. Classes with diverse images could have lower thresholds, while highly consistent crops could demand higher similarity. The system would stay robust even if the dataset was imbalanced.
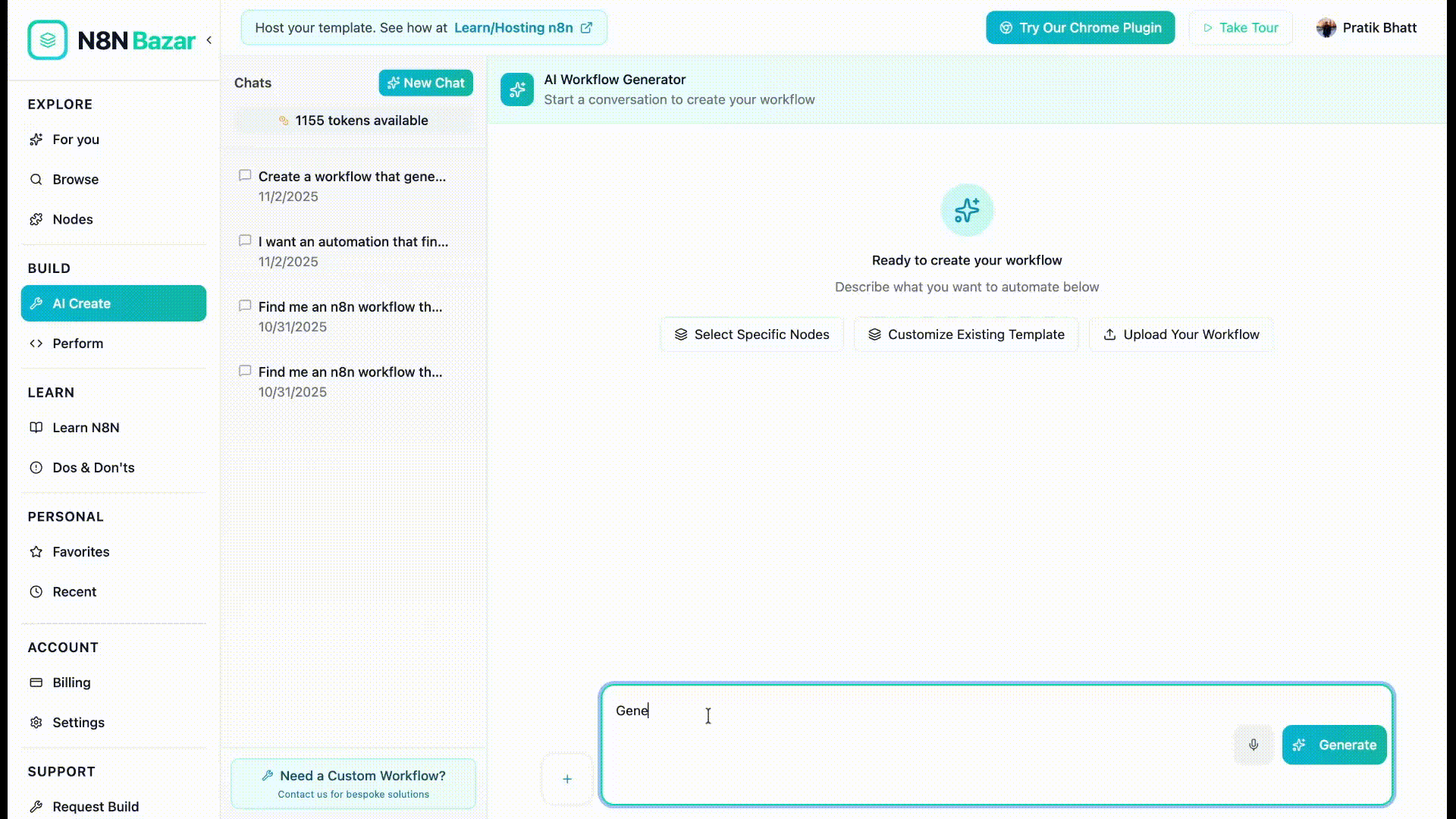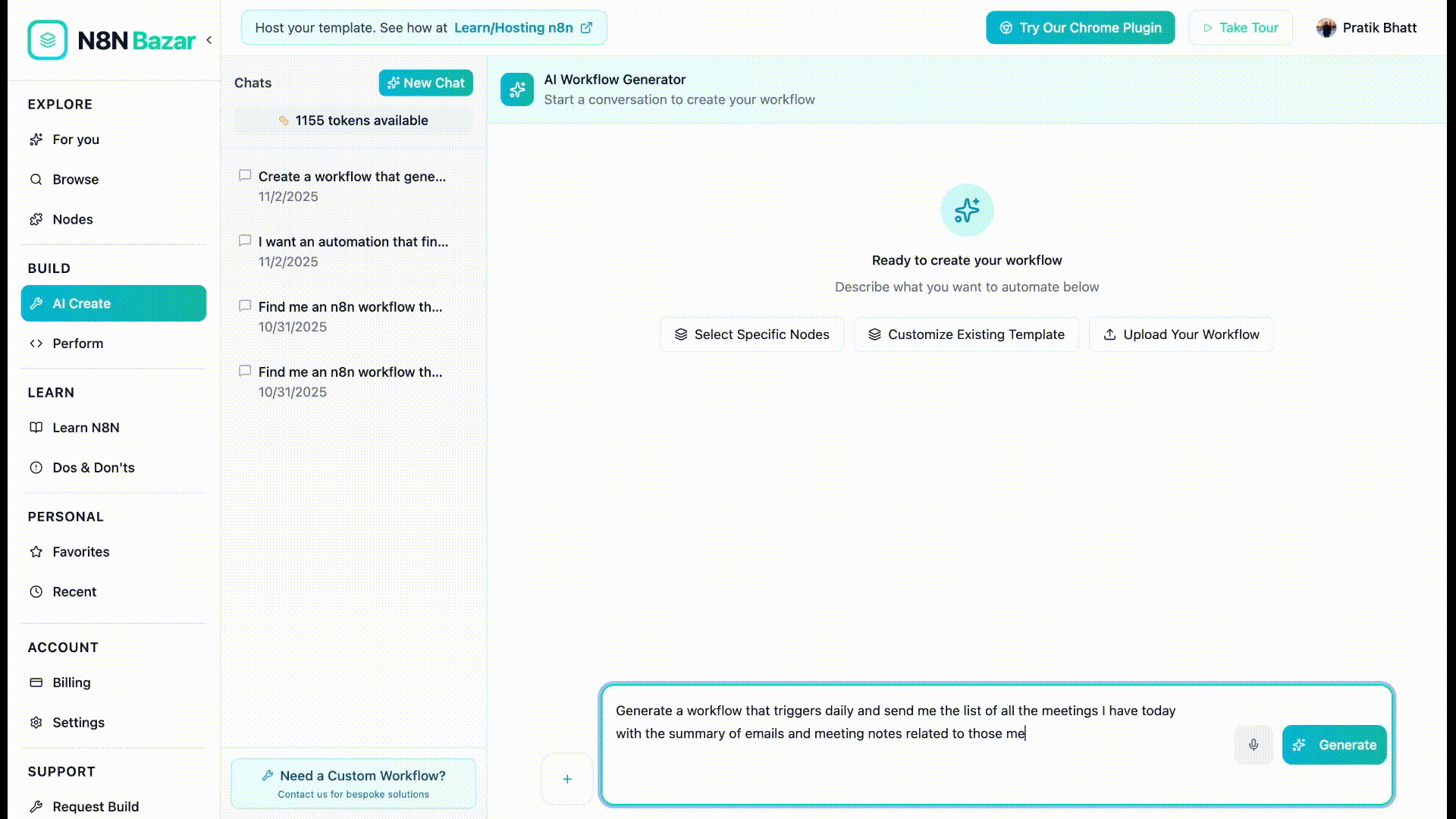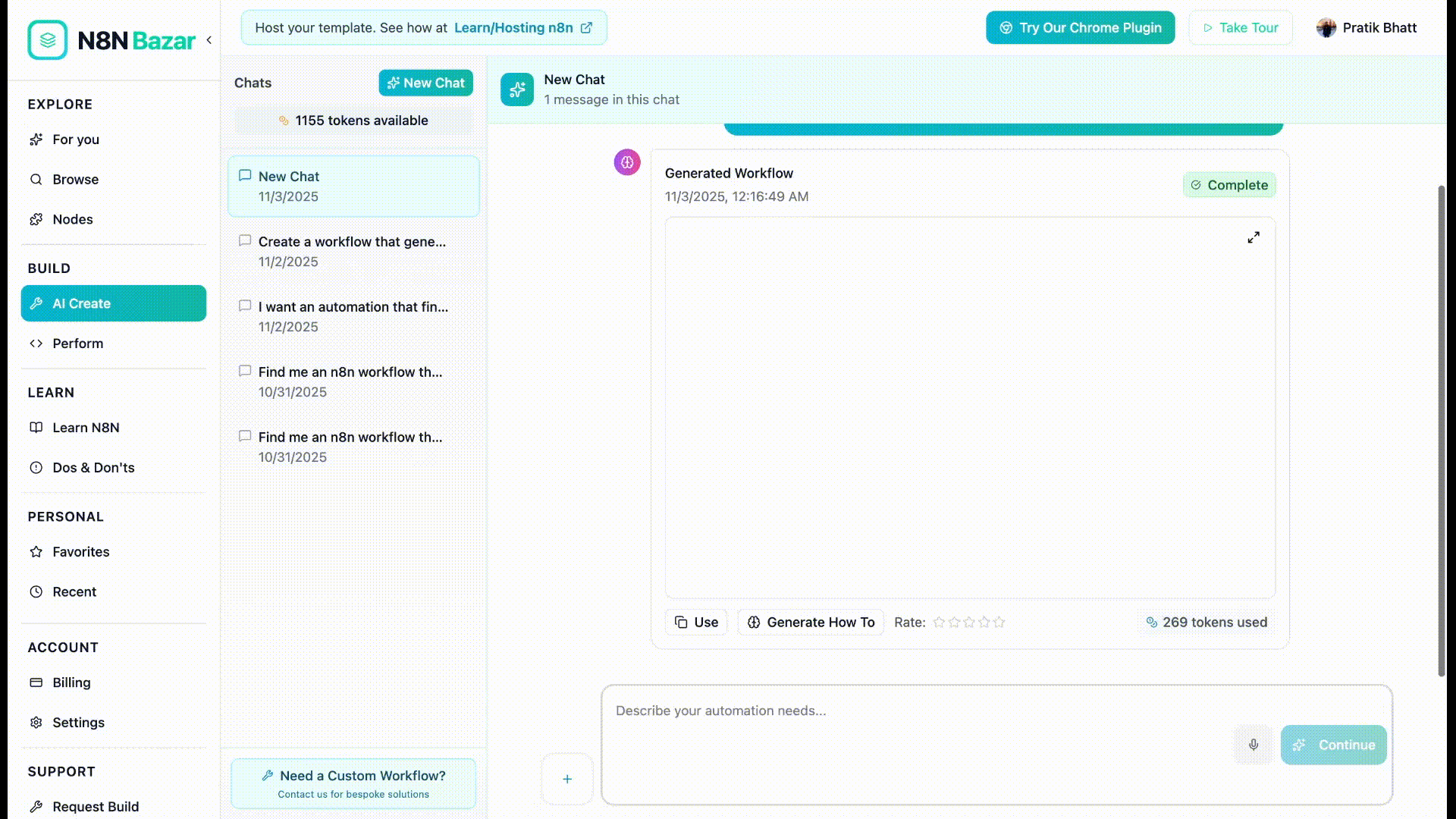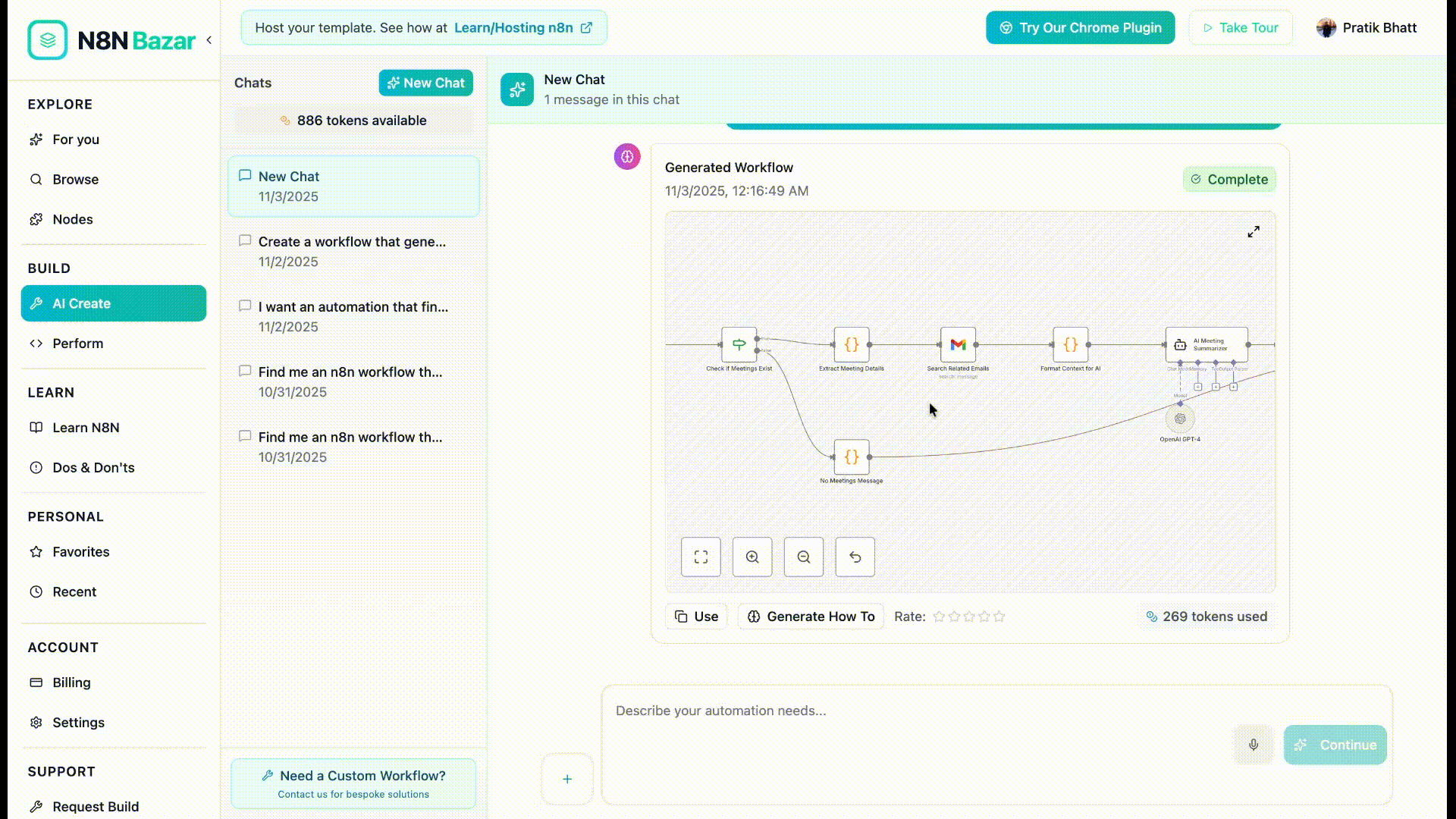
The Turning Point: Wiring It All Together in an n8n Workflow
With Qdrant populated and medoids defined, Priya turned to the heart of the system: an n8n workflow template that would take any new image URL and decide if it belonged to a known crop or if it was anomalous.
How the n8n pipeline flows
Instead of a tangled mess of scripts, the workflow was clean and modular. Each node had a clear role.
- Execute Workflow Trigger – The entry point. It receives the incoming image URL from another workflow, a webhook, or a scheduled job.
- Set (Image URL hardcode) – Normalizes or hardcodes the image URL so downstream nodes always receive it in a consistent format.
- Voyage Embeddings Request – Calls the Voyage AI multimodal embeddings API and transforms the image into an embedding vector suitable for vector search.
- Qdrant Query – Sends the embedding to the Qdrant collection, requesting only points marked with
is_medoid == true. This keeps queries efficient and focused on cluster centers. - Compare Scores (Code) – A small but critical code node. It inspects each returned medoid, compares its similarity score with the stored
is_medoid_cluster_threshold, and decides whether the image is anomalous.
Helper nodes supported this core logic by:
- Counting how many crop classes existed in the collection.
- Setting the Qdrant query limit to match the number of medoids.
The decision logic that makes or breaks anomaly detection
In the Compare Scores node, Priya implemented the logic that would finally solve her original problem.
For each query:
- Qdrant returns a set of medoids with their similarity scores to the input image.
- The code node checks, for every medoid:
- If
similarity_score >= is_medoid_cluster_threshold, the image is considered a match for that cluster.
- If
- If at least one medoid passes its threshold, the workflow:
- Marks the image as not anomalous.
- Reports the best matching crop class.
- If no medoid meets its threshold, the workflow:
- Flags the image as a potential new or undefined crop.
- Emits an alert for follow up by agronomists or data scientists.
In human readable form, the Compare Scores node returns one of two messages:
"Looks similar to {crop_name}"– when at least one medoid’s score is above its threshold."ALERT, we might have a new undefined crop!"– when none of the medoids meet their thresholds.
For Priya, this was the moment the system became truly useful. The workflow did not just output numbers. It gave clear, actionable decisions.
Deploying the Workflow: From Prototype to Production
Once the logic was tested on a few sample images, Priya needed to make it reliable and secure enough for daily use across farms.
Credentials and configuration
She configured the following in n8n:
- Voyage AI API key using HTTP header authentication for the embeddings request node.
- Qdrant Cloud credentials including the collection name and cloud URL.
- Environment variables and n8n credential stores so that no secrets were hardcoded in the workflow.
With these in place, deploying the anomaly detection workflow was as simple as:
- Pointing the workflow to the correct Qdrant collection.
- Ensuring the medoid payload fields (
is_medoidandis_medoid_cluster_threshold) matched the configuration. - Connecting the workflow to upstream systems that provided image URLs, such as field apps or internal labeling tools.
Security and privacy in the field
Because the system processed real farm images, Priya enforced a few rules:
- Only pass image URLs that the team was authorized to process.
- Keep all API keys and credentials secure in n8n’s credential store or environment variables.
- Restrict access to the workflow to trusted services and internal users.
Living With the Workflow: Troubleshooting and Tuning
Once farmers started sending thousands of images through the system, some patterns emerged. The workflow worked, but there were edge cases. Priya used a few key strategies to tune performance.
Common issues and how she fixed them
- Low similarity scores for almost every image
When embeddings looked uniformly low, she verified that:- The same Voyage AI model and version were used for both the original collection and the live queries.
- No embedding drift had occurred due to a silent model upgrade.
- Too many false positives (normal images flagged as anomalies)
Priya:- Analyzed the similarity score distributions per cluster.
- Adjusted medoid thresholds upward where needed.
- Used cluster-wise ROC curves to find better thresholds per crop class.
- Missing or misidentified medoids
When some classes never matched, she checked that:- Medoids had been correctly flagged with
is_medoid == true. - The
is_medoid_cluster_thresholdfield was present and correctly named.
- Medoids had been correctly flagged with
- Scaling up
As the dataset grew, she:- Kept queries efficient by always filtering on
is_medoid == true. - Relied on Qdrant’s ability to scale a single collection while keeping medoid-only queries fast.
- Kept queries efficient by always filtering on
Measuring Success: Evaluation and Monitoring in Production
Priya knew that an anomaly detector is only as good as its long term behavior. To keep confidence high, she set up a simple evaluation and monitoring loop.
- Validation set evaluation
She used a held-out set of labeled images, including known anomalies, to:- Measure detection accuracy.
- Track true positives and false negatives for anomalies.
- Monitoring similarity distributions
Over time, she:- Tracked the distribution of medoid similarity scores.
- Set alerts when distributions shifted, a sign of concept drift or changing field conditions.
- Human review of flagged anomalies
For a sample of flagged images, domain experts:- Reviewed whether they were truly unknown crops.
- Identified misclassifications or mislabeled data.
- Fed confirmed new examples back into the dataset for future model updates.
What This Enabled: Real Use Cases on Real Farms
Once in place, the n8n, Qdrant and Voyage AI workflow changed how Priya’s team handled incoming images.
- Field data capture
Photos from scout walks were automatically checked against known farm crops. Anything unfamiliar was flagged for agronomists to review. - Quality control for datasets
Before training new models, the pipeline scanned image datasets to detect mislabeled or corrupted images. - Species discovery and rare events
Images that did not match any crop class surfaced possible new species, invasive weeds, or rare disease patterns that were not in the original dataset. - Multi modal extensions
Where contextual metadata or text descriptions were available, Priya explored combining image and text embeddings to improve detection accuracy.
What had started as a problem of noisy labels and unknown plants turned into a flexible, extensible anomaly detection framework.
The Resolution: From Red Alerts to Reliable Insights
Weeks after deploying the workflow, the red alerts on Priya’s dashboard looked different. They were fewer, clearer, and far more meaningful.
Instead of random spikes from mislabeled data, the system highlighted images that truly did not belong to any known crop cluster. Some were new pest outbreaks. Others were weeds that had crept into fields. A few revealed labeling mistakes that would have poisoned future training runs.
Most importantly, the workflow was explainable. For every decision, Priya could see:
- Which medoids were closest.
- What the similarity scores were.
- How those scores compared to per cluster thresholds.
It was no longer a black box model. It was a transparent, data driven pipeline that her team could trust, tune, and extend.
Try the Same n8n Template in Your Own Stack
If you already have a Qdrant collection and embeddings, you can plug this n8n crop anomaly detection workflow into your environment in minutes and start flagging unknown crops or anomalous images.
To reproduce Priya’s setup:
- Prepare a crops dataset and upload images to a cloud bucket.
- Create a Qdrant collection and upload image embeddings and metadata.
- Run a medoids setup pipeline to compute medoids and per cluster thresholds, setting
is_medoidandis_medoid_cluster_thresholdpayload fields. - Configure n8n credentials for Voyage AI and Qdrant securely.
- Deploy the anomaly detection workflow and point it to your Qdrant collection and cloud URL.
From there, you can:
- Run the workflow on a sample image.
- Tune per cluster thresholds using a validation set.